Oddly enough, one of the outstanding yet fundamental questions in the study of biodiversity remains “how many species are there?” This may come as a surprise to many, but the fact remains that the actual number of eukaryotic species currently described is ~1.4–1.9 million with estimates on the actual number ranging from 9 million to much higher depending on the source. Even this modest prediction suggests that we have documented only ~18% of eukaryotic life or one in five of the eukaryotic species on the planet.
Growing up as a teen on the South Shore of Nova Scotia my life was centered on the ocean and a curiosity of the species that called it home. Fortunately that childhood passion has transitioned into a research career that is focused on a particular lineage of life, the red algae. Through our research, my colleagues and I attempt to determine the number of species of red algae, their respective relationships to one another, and their distribution in the world’s oceans. This is no trivial task because red algae are notoriously difficult to identify.
Red algae have simple morphologies that, through a process termed convergent evolution, can result in distantly related species looking more similar to one another than they do to their respective closer relatives. Consider how bats and birds both have wings but bats and wingless humans are more closely related to each other than bats are to birds.

Top left and right, respectively, are our recently described Rhodymenia norfolkensis and an unnamed species of Schottera—impossible to tell apart while collecting in Australia, but from different orders of life. On the bottom are two of the many different morphologies of the phenotypically plastic Chondracanthus harveyanus from British Columbia.
Most of our taxonomic advances have resulted from DNA barcoding surveys of marine algae. DNA barcoding is basically the application of standardized molecular markers to assign biological specimens to species. The premise is simple enough—individuals of the same species have similar DNA relative to those of other species. By reading DNA through a process termed DNA sequencing, a specimen, or any form of biological material (example, a dried powder sold in a market as Irish Moss—a type of seaweed), can be matched to its species. To assign unknown biological material to a known species, however, DNA sequences from that species must have been previously included in a DNA barcoding library to facilitate a match. It has been through the process of building this reference library that DNA barcoding has exposed a wealth of undocumented species. New species are discovered when analyzing samples from wide-scale collections in places that might otherwise not have been visited and when DNA barcoding indicates that there are two or more species where we thought that there was only one.Conversely, red algae are highly prone to phenotypic plasticity; that is their morphology can vary in different environments such that individuals within a species can look more different to one another than some of them do to individuals of another species. Obviously, this can make it difficult to establish the number of red algal species.
What is fun about our latest discovery of a new order of red algae and its species Corynodactylus rejiciendus is that the DNA barcode was read accidentally. We were attempting to generate sequence data to include the red alga Camontagnea oxyclada in the reference barcode library, but recovered a DNA sequence for an unknown red algal species. The unknown species turned out to be one of the many red microalgae that live in and on other marine algae and animals. We have long known that there are microscopic red (and green and brown) algae that live among the cells of larger red, green, and brown seaweeds as well as within marine animals. However, these microscopic red algae are notoriously understudied, and little is known of their richness, distribution, and evolutionary diversity. The discovery here is one in a series of similar recent accidents that have opened our eyes to this poorly explored window of red algal diversity. Although the discovery here of Corynodactylus rejiciendus was initially accidental, we used contemporary molecular methods to explore further this tiny red alga to establish that it represented an entirely new order of life.

Tiny (<100µm) plants of Corynodactylus rejiciendus smothering the host Camontagnea oxyclada. Corynodactylus loosely means club-shaped finger referring to the shape of the erect filaments while rejiciendus means fit to be rejected or thrown out, which was the original fate of the “incorrect” DNA barcode sequence.
In some cases cryptic species are hidden in plain sight in areas that have been heavily studied by algal taxonomists. For example, the seaweeds Ptilota haidarum and Schizymenia tenuiseluded detection despite decades of intense taxonomic study along the coasts of British Columbia; they were masked by the similar looking species Ptilota hypnoides and Schizymenia pacifica, respectively.So, how many species of red algae are there? Currently there are ~7000 named species, but estimates run as high as 14,000 total species. However, it is clear that many red algae (and other host species) have other red algal species growing in or on them and that much of this diversity remains poorly documented by science. I would speculate that we have as many as 20,000 species of red algae. For our part my colleagues and I have added about 130 species to this tally over the past few decades, but this represents only a fraction of those that we have discovered—assigning a name to a species can be a complicated task so most of these species are awaiting formal publication.

Ptilota hypnoides (left) and its cryptic cousin Ptilota haidarum (right) growing together in the pristine waters of Haida Gwaii.
In other cases some of these species are completely new to science having been discovered in parts of the world that have been underexplored. For example, a trip to the Cocos (Keeling) Islands, Australia, in the eastern Indian Ocean has resulted in dozens of new red algal species that we are just starting to formally publish—only four have been described to date, Incendia undulata, Leiomenia imbricata, Meredithia compaginata, and Verlaquea fimbriata.

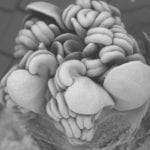
Newly described from the Cocos Keeling Islands, Meredithia compaginata starts as little cup-shaped blades that eventually bear further cups from the margins to form a complex, albeit tiny, alga.
To date we have accidentally uncovered seven other new species in the way that we uncovered Corynodactylus rejiciendus, despite trying to avoid these accidents in the first place; we are after all aiming for the DNA barcode sequence from the host algal specimen not the microalgae living in/on it. However, the availability of new sequencing technologies will significantly increase our ability as scientists to explore directly the richness of microscopic red algae in an endless variety of hosts.Some areas are so understudied, yet so rich in their red algal flora, that we still find new families and orders of life such as the family Acrothesauraceae and order Entwisleiales from the beautiful waters of Tasmania, Australia. To put this into context, the mammalian order Primates includes species as diverse as lemurs, monkeys, and humans. To discover a new order of life is by most measures remarkable, but not uncommon for the red algae as our current study attests. Add to the previous discoveries the undocumented wealth of microscopic red algae living in and on those species and it is clear that the study of diversity among red algae is still an open book.

Picture of the author taken by a long-time colleague Kyatt Dixon as they worked the flora of Norfolk Island in the South Pacific.